Lempradl
Laboratory
Intergenerational Inheritance of Nutritional States
 The laboratory of Dr. Adelheid (Heidi) Lempradl investigates how metabolism shapes developmental trajectories, acting as both a driver of early developmental processes and a determinant of disease later in life. This dual focus guides the Lempradl Lab’s research program, which is both mechanistically rigorous and directly relevant to public health. Early disruptions to metabolic programming are increasingly recognized as root causes of chronic conditions, including metabolic and neurodevelopmental disorders. The strategic investment in innovative multi-omic methodologies allows the Lempradl Lab to define how metabolism unfolds during normal early development and examine how reprogramming occurs following environmental challenges.
The laboratory of Dr. Adelheid (Heidi) Lempradl investigates how metabolism shapes developmental trajectories, acting as both a driver of early developmental processes and a determinant of disease later in life. This dual focus guides the Lempradl Lab’s research program, which is both mechanistically rigorous and directly relevant to public health. Early disruptions to metabolic programming are increasingly recognized as root causes of chronic conditions, including metabolic and neurodevelopmental disorders. The strategic investment in innovative multi-omic methodologies allows the Lempradl Lab to define how metabolism unfolds during normal early development and examine how reprogramming occurs following environmental challenges.
The Lempradl Lab strives to provide an environment that fosters independence, curiosity, and resilience and to prepare trainees not just for experiments, but for scientific careers.
Lempradl Lab Values: Teamwork, Innovation, Reliability, Wellbeing, Respect
Join Our Teams
The labs of Dr. Adelheid Lempradl and Dr. Andrew Pospisilik are seeking to hire a Postdoctoral Fellow or Research Scientist for a combined project focused on exploring two unique and powerful models of paternal epigenetic inheritance.
Recent News & Press
Learn More
A clearer view: How a commonly studied gene impacts more than eye color in fruit fly research

Study offers high-resolution look at ‘metabolic handoff’ from fruit fly mothers to embryos

Van Andel Institute, The Scientist launch podcast mini-series on health and the environment
OUR IMPACT
We’re raising thousands to save millions.
We’re turning hope into action for the millions of people around the world affected by diseases like cancer and Parkinson’s. Find out how you can help us make a difference.
- 122 peer-reviewed papers published in 2024, 63 of which were in high-impact journals
- 15 VAI-SU2C Epigenetics Dream Team clinical trials launched to date
- 10 clinical trials co-funded by VAI & Cure Parkinson's (out of 41 total International Linked Clinical Trials Program trials)
Adelheid (Heidi) Lempradl, Ph.D.
Associate Professor, Department of Metabolism and Nutritional Programming
Areas of Expertise
Epigenetics, metabolism, intergenerational inheritance, nutritional programming
Biography
Dr. Adelheid (Heidi) Lempradl is an associate professor in the Department of Metabolism and Nutritional Programming at Van Andel Institute. Her research program advances our fundamental understanding of how early life shapes lifelong physiology and disease risk.
Dr. Lempradl uses an integrative approach that combines developmental biology, metabolism, environmental health, genetics and innovative multi-omic technologies. She pioneered a single-embryo multi-omics platform that reveals, at high resolution, how transcriptional and metabolic programs unfold in early embryos and how they are altered by parental diet or environmental exposures. Her work aims to define the molecular mechanisms that mediate intergenerational metabolic effects and to identify pathways that contribute to disease susceptibility.
Dr. Lempradl earned her Ph.D. at the University of Vienna. She completed postdoctoral training at the Institute of Molecular Biotechnology in Austria and the Max Planck Institute of Immunobiology and Epigenetics in Germany, where her work contributed to a growing understanding of how environmental exposure can leave molecular imprints that persist beyond a single generation. At VAI, she leads a growing research program and is an active mentor and educator within the Van Andel Institute Graduate School.
Selected Publications
Pérez-Mojica JE, Madaj ZB, Isaguirre C, Roy J, Lau KH, Sheldon RD, Lempradl A. 2025. Resolving early embryonic metabolism in Drosophila through single-embryo metabolomics and transcriptomics. Nat Metab.
*Featured on the cover
Rickle A, Sudhakar K, Booms A, Stirtz E, Lempradl A. 2025. More than meets the eye: Mutation of the white gene in Drosophila has broad phenotypic and transcriptomic effects. Genetics.
Dror E, Fagnocchi L, Wegert V, Apostle S, Grimaldi B, Gruber T, Panzeri I, Heyne S, Höffler KD, Kreiner V, Ching R, Lu TTH, Semwal A, Johnson B, Senapati P, Lempradl A, Schones D, Imhof A, Shen H, Pospisilik JA. 2023. Epigenetic dosage identifies two major and functionally distinct β cell subtypes. Cell Metab.
Pérez-Mojica JE, Enders L, Walsh J, Lau KH, Lempradl A. 2023. Continuous transcriptome analysis reveals novel patterns of early gene expression in Drosophila embryos. Cell Genom.
Yang CH, Longinotto J, Panzeri I, Arrigoni L, Wegert V, Heyne S, Seifert G, Lempradl A, Bonisch U, Pospisilik JA. 2022. Isolation and processing of murine white adipocytes for transcriptome and epigenome analyses. J Vis Exp 184:e64128.
Yang CH*, Fagnocchi L*, Apostle S, Wegert V, Casani-Galdón S, Landgraf K, Panzeri I, Dror E, Heyne S, Wörpel T, Chandler DP, Lu D, Yang T, Gibbons E, Guerreiro R, Brás J, Thomasen M, Grunnert LG, Vaag AA, Gillberg L, Grundberg, E, Conesa A, Körner A, PERMUTE, Pospisilik JA. 2022. Independent phenotypic plasticity axes define distinct obesity subtypes. Nat Metab.
*Co-first authorship
**Highlighted in News & Views
***Dr. Lempradl is part of PERMUTE
Lempradl A. 2019. Germ cell-mediated mechanisms of epigenetic inheritance. Semin Cell Dev Biol.
Justice AE…Lempradl A…Lindgren CM. 2019. Protein-coding variants implicate novel genes related to lipid homeostasis contributing to body-fat distribution. Nat Genet 51(3):452-469.
Lu TT, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang CH, Sagar, Arrigoni L, Dalgaard K, Terperino R, Enders L, Selveraj M, Ruf M, Raja SJ, Xie H, Boenisch U, Orkin Sh, Lynn FC, Hoffman BG, Grün D, Vovouri T, Lempradl A, Pospisilik JA. 2018. The Polycomb-dependent epigenome controls b cell dysfunction, dedifferentiation and diabetes. Cell Metab 27(6):1294–1308.
Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M, Hendricks AE, Karaderi T, Lempradl A, …, Pospisilik JA, Rivadeneira F, Borecki IB, Deloukas P, Frayling TM, Lettre G, North KE, Lindgren CM, JN Hirschhorn, Loos RJF. 2018. Protein-altering variants associated with body mass index implicate 1 pathways that control energy intake and expenditure underpinning obesity. Nat Gen 50(1):26–41.
Theurich S, Tsaousidou E, Hanssen R, Lempradl A, Mauer J, Timper K, Schilbach K, Folz-Donahue K, Heilinger C, Sexl V, Pospisilik JA, Wunderlich FT, Brüning JC. 2017. IL-6/Stat3-dependent induction of a distinct, obesity-associated NK cell subpopulation deteriorates energy and glucose homeostasis. Cell Metab 26(1):171–184.
Dalgaard K, Landgraf K, Heyne S, Lempradl A, Longinotto J, Gossens K, Ruf M, Orthofer M, Strogantsev R, Selvaraj M, Casas E, Teperino R, Surani MA, Zvetkova I, Rimmington D, Tung L, Larder R, Yeo GSH, O’Rahilly S, Whitelaw E, Penninger JM, Jenuwein T, Cheung CL, Ferguson-Smith AC, Coll AP, Körner A, Pospisilik JA. 2016. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell 164(3):353–364.
Lempradl A, Penninger JM, Pospisilik JA. 2015. Exploring the emerging complexity in transcriptional regulation of energy homeostasis. Nat Rev Genet 16(11):665–681.
Ost A*, Lempradl A*, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U,Itskov PM, Stoeckius M, Ruf M, Rajewsky N, Reuter G, Iovino N, Ribeiro C, Alenius M, Heyne S, Vavouri T, Pospisilik JA. 2014. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159(6):1352–1364.
*Co-first authors
Herzog VA*, Lempradl A*, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, Ruf M, Pospisilik A, Dimond A, Senergin HB, Vargas ML, Simon JA, Ringrose L. 2014. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet 46(9):973–981.
*Co-first authors
Teperino R, Lempradl A, Pospisilik JA. 2013. Bridging epigenomics and complex disease: the basics. Cell Mol Life Sci 70(9):1609–1621.
Lempradl A, Ringrose L. 2008. How does noncoding transcription regulate Hox genes? Bioessays 30(2):110–121.
Stampfl S, Lempradl A, Koehler G, Schroeder R. 2007. Monovalent ion dependence of neomycin B binding to an RNA aptamer characterized by spectroscopic methods. Chembiochem 8(10):1137–1145.
Lempradl A, Stampfl S, Koehler G. 2004. RNA-antibiotics interactions: A spectroscopic study of conformational changes upon binding. 48th Annual Meeting of the Biophysical Society, Baltimore Biophysical Journal 86(1):143A–143A part 2.
Projects
Early development is a uniquely dynamic period, during which cell fates are determined, organs form, and plasticity is at its peak. Our work is guided by the idea that metabolism is not merely a passive supplier of energy and building blocks, but an active, instructive layer of developmental regulation. By mapping when and how key metabolic transitions unfold, and how they are perturbed by environmental exposures, we aim to identify the mechanisms through which developmental trajectories are established and modified.
There are three major project areas in the lab:
Projects one & two
Developmental programming by maternal nutrition: How does maternal obesity lead to embryonic reprogramming?
The conceptual foundation for this work draws on earlier studies by Dr. Lempradl and others demonstrating that the parental environment, particularly nutrition, can influence offspring phenotype in the absence of direct genetic changes. Building on this, the lab became interested in how the maternal metabolic environment, specifically obesity, might precondition the embryo through oocyte-derived cues.
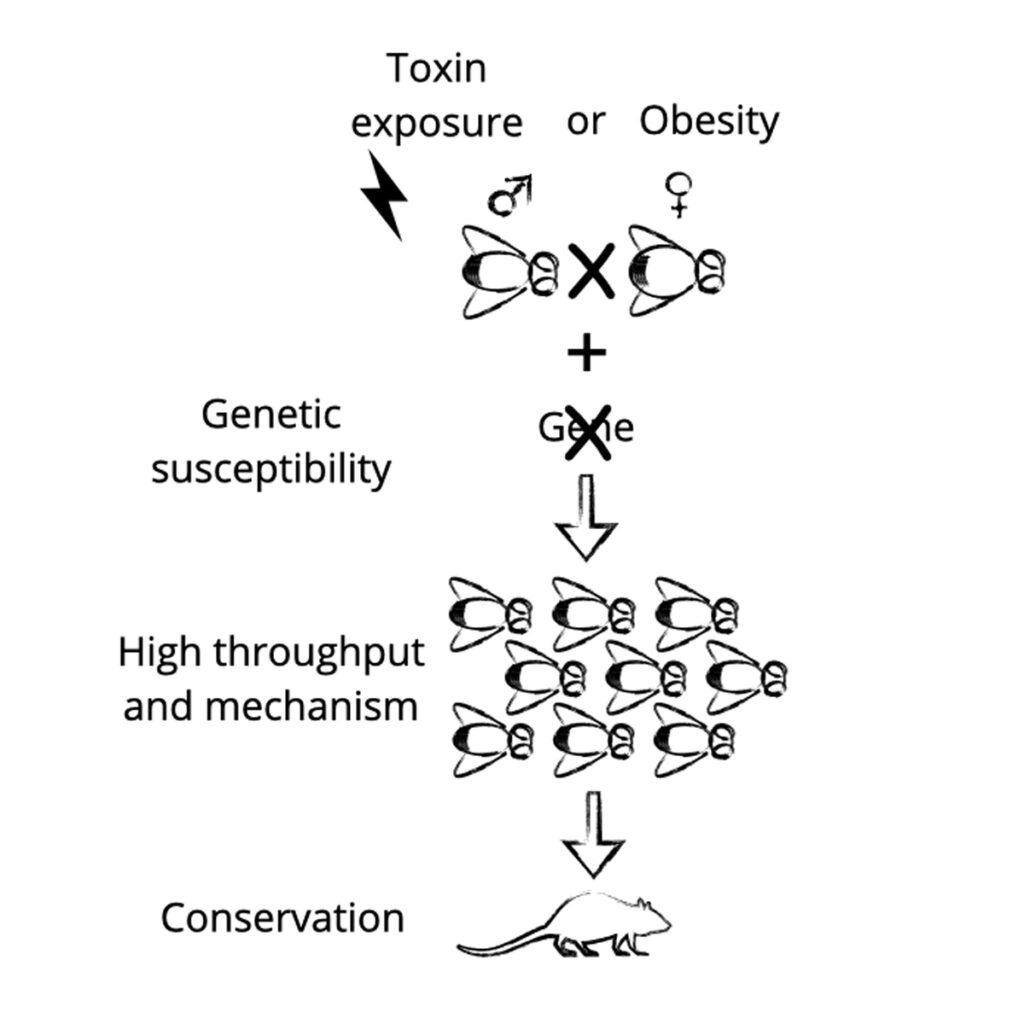 To examine how maternal obesity alters the metabolic and transcriptomic programming of the oocyte and early embryo, we apply our single-embryo multi-omics approach. Additionally, the lab investigates how these cues might interact with genetic predisposition to disease. By identifying modifiable pathways that mediate maternal effects on development, this research opens the door to translational strategies aimed at prevention or mitigation, particularly in the context of rising global rates of maternal obesity and metabolic dysfunction.
To examine how maternal obesity alters the metabolic and transcriptomic programming of the oocyte and early embryo, we apply our single-embryo multi-omics approach. Additionally, the lab investigates how these cues might interact with genetic predisposition to disease. By identifying modifiable pathways that mediate maternal effects on development, this research opens the door to translational strategies aimed at prevention or mitigation, particularly in the context of rising global rates of maternal obesity and metabolic dysfunction.
Environmental toxicants and paternal programming: How do environmental toxicants alter developmental programming and metabolic health across generations?
This project addresses a critical gap by investigating how environmental exposures, particularly through the paternal germline, alter developmental programming in ways that predispose offspring to disease. Our integrative approach combines high-throughput discovery and mechanistic analysis in Drosophila, where genetic tools and rapid generation times enable detailed interrogation of causal pathways, and complements these studies with targeted validation in mouse models. This provides a powerful and cost-effective platform to identify, prioritize and characterize environmental exposures with intergenerational consequences.
Our goal is to identify transcriptional signatures and disrupted developmental pathways in embryos derived from exposed fathers, with a focus on genes linked to metabolic regulation, epigenetic remodeling and stress responses. These data will be integrated with small RNA and sperm epigenome profiling from the same exposure model, providing a comprehensive view of how paternal exposure reprograms embryonic development at multiple molecular levels. Longer term, we aim to expand this approach to other toxicants identified in our high throughput Drosophila model and further refine cross-species comparisons of conserved early-response pathways.
Project three
High-resolution view of developmental metabolism: Developing the tools and baselines needed to understand metabolism in early development.
 A defining feature of my lab has been our focus on developing the tools needed to understand metabolism in early development, a biologically complex window that is difficult to access experimentally. The investment in building these methods is strategic: it allows us to define how metabolism unfolds during normal early development and study reprogramming in the project areas described above. We recently established a single-embryo multi-omics platform that combines RNA-sequencing and metabolomics in individual Drosophila embryos. This system was designed to treat development as a continuous molecular process rather than a series of morphologically defined stages. By pairing transcriptomics and metabolomics in the same embryo, we not only provide descriptive maps but also generate mechanistically informed hypotheses about how metabolites act as signals that shape developmental trajectories.
A defining feature of my lab has been our focus on developing the tools needed to understand metabolism in early development, a biologically complex window that is difficult to access experimentally. The investment in building these methods is strategic: it allows us to define how metabolism unfolds during normal early development and study reprogramming in the project areas described above. We recently established a single-embryo multi-omics platform that combines RNA-sequencing and metabolomics in individual Drosophila embryos. This system was designed to treat development as a continuous molecular process rather than a series of morphologically defined stages. By pairing transcriptomics and metabolomics in the same embryo, we not only provide descriptive maps but also generate mechanistically informed hypotheses about how metabolites act as signals that shape developmental trajectories.
Beyond its technical value, this work advances a conceptual shift in how metabolism is viewed in development. Traditionally seen as supportive, metabolism is emerging as an active organizer of embryogenesis. As interest grows in the link between early metabolism, environmental exposures and disease susceptibility, advancing our technology platform ensures that the lab remains positioned at the forefront of both methodological and conceptual innovation. The tools and datasets we have built not only support our own research trajectory but offer broad utility across model systems and disciplines, opening new avenues for mechanistic discover and translational impact.


Dmitri Martirosov
Laboratory Aide, Department of Metabolism and Nutritional Programming

Eduardo Perez-Mojica, Ph.D.
Postdoctoral Fellow, Lempradl Laboratory
Mechanistic dissection of epigenetic inheritance

Joe Roy, BLS, B.S.
Assistant Research Technician, Department of Metabolism and Nutritional Programming

April Rickle
Ph.D. Student, VAI Graduate School
Thesis: Impact of toxins and nutrients on brain function and development

Ellen Stirtz, B.S.
Ph.D. Student, VAI Graduate School
Thesis: The combined effect of maternal obesity and autism spectrum disorder-associated genetic mutations on neurodevelopment.

Krittika Sudhakar, Ph.D.
Postdoctoral Fellow, Lempradl Laboratory
Thesis: The effect of maternal nutrition on offspring development and metabolism

Jeanie Wedberg
Senior Administrative Assistant II


